Download HarvestChoice indicators in raster and/or tabular formats
Package results from getLayer into tabular or spatial raster format.
Also includes README and Tabular Data Package specifications.
Currently supported export formats include CSV (csv), STATA (dta), GeoJSON (geojson),
GeoTIFF (tif), R raster (grd), RData (rda), ESRI ASCII raster (asc), and netCDF (nc).
Calling genFile(var="bmi", iso3="TZA", format="dta") is equivalent to calling
the convenience method hcapi(var="bmi", iso3="TZA", format="dta").
genFile(var, iso3 = "SSA", by = NULL, format = c("csv", "geojson", "tif", "dta", "asc", "rds", "grd"), dir = ".", ...)
Arguments
| var | character array of indicator codes, passed to |
|---|---|
| iso3 | character array of ISO3 country or region codes, passed to |
| by | character array of indicator codes to summarize by, passed to |
| format | output format, one of "csv", "json", "tif", "dta", "asc", "grd", "rds". |
| dir | output directory, default to current working directory |
| ... | any other optional argument passed to |
Value
character, array of generated file names included in the data package
Details
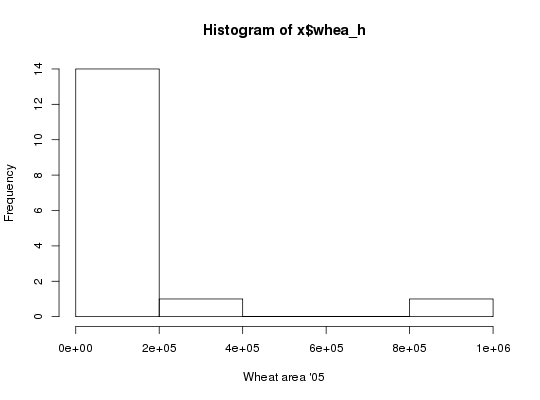
API call: total wheat harvested area across 16 agro-ecological zones in Nigeria and
Ethiopia in STATA format
$ curl http://hcapi.harvestchoice.org/ocpu/library/hcapi3/R/genFile \
-d '{"var" : "whea_h", "iso3" : ["NGA", "ETH"], "by" : "AEZ16_CLAS", "format" : "dta"}' \
-X POST -H 'Content-Type:application/json'
/ocpu/tmp/x0e654538b7/R/.val
/ocpu/tmp/x0e654538b7/stdout
/ocpu/tmp/x0e654538b7/warnings
/ocpu/tmp/x0e654538b7/source
/ocpu/tmp/x0e654538b7/console
/ocpu/tmp/x0e654538b7/info
/ocpu/tmp/x0e654538b7/files/DESCRIPTION
/ocpu/tmp/x0e654538b7/files/README
/ocpu/tmp/x0e654538b7/files/whea_h-AEZ16_CLAS-NGA.dta
GET all generated files in a ZIP archive
$ wget http://hcapi.harvestchoice.org/ocpu/tmp/x0e654538b7/zip
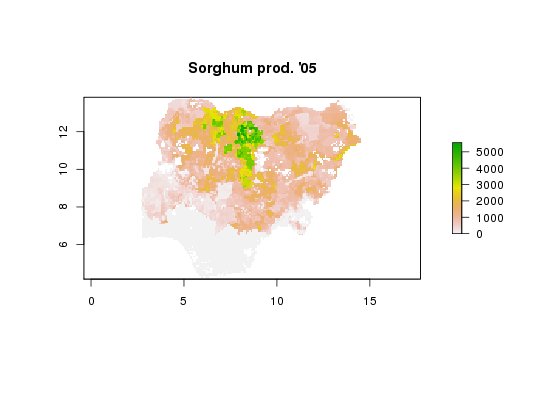
API call: sorghum production in Nigeria in ESRI ASCII raster format
$ curl http://hcapi.harvestchoice.org/ocpu/library/hcapi3/R/genFile \
-d '{"var" : "sorg_p", "format" : "asc"}' \
-X POST -H "Content-Type:application/json"
--
/ocpu/tmp/x02a7a044c7/R/.val
/ocpu/tmp/x02a7a044c7/stdout
/ocpu/tmp/x02a7a044c7/warnings
/ocpu/tmp/x02a7a044c7/source
/ocpu/tmp/x02a7a044c7/console
/ocpu/tmp/x02a7a044c7/info
/ocpu/tmp/x02a7a044c7/files/DESCRIPTION
/ocpu/tmp/x02a7a044c7/files/README
/ocpu/tmp/x02a7a044c7/files/sorg_p--SSA.asc
/ocpu/tmp/x02a7a044c7/files/sorg_p--SSA.asc.aux.xml
/ocpu/tmp/x02a7a044c7/files/sorg_p--SSA.prj
GET all generated files in a ZIP archive
$ wget http://hcapi.harvestchoice.org/ocpu/tmp/x02a7a044c7/zip
See also
datapackage to generate associated metadata records
Examples
# Total wheat harvested area across 16 agro-ecological zones in Nigeria and Ethiopia # in STATA format x <- genFile("whea_h", iso3=c("NGA", "ETH"), by="AEZ16_CLAS", format="dta")#> Warning: the condition has length > 1 and only the first element will be used#> Warning: the condition has length > 1 and only the first element will be used# Load generated STATA file require(foreign)#>x <- read.dta(x[1]) # Plot histogram with full layer title hist(x$whea_h, xlab=vi["whea_h", varLabel])# Sorghum production in Nigeria in ESRI ASCII raster format x <- genFile("sorg_p", iso3="NGA", format="asc")#> Warning: colorTables valid for Byte type only in some drivers#> Warning: Unable to set color tablex#> [1] "./hcapi-nga-sorg_p.asc" "./hcapi-nga-sorg_p.asc.aux.xml" #> [3] "./hcapi-nga-sorg_p.prj" "./meta.csv" #> [5] "./README.md" "./datapackage.json"# Load and plot generated raster require(raster)#>#>#> #>#> #> #>x <- raster(x[1]) plot(x, main=vi["sorg_p", varLabel])cellStats(x, "mean")#> [1] 897.064